Clustering method¶
This tutorial gives a bit more detail about clustering methods and how to implement your own.
# Prerequisites
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams["figure.dpi"] = 300
plt.rcParams["figure.figsize"] = (12, 5)
# Include this once we have a published release to fetch test data
# from toad.utils import download_test_data
# download_test_data()
from toad import TOAD
from toad.shifts import ASDETECT
td = TOAD("test_data/garbe_2020_antarctica.nc", time_dim="GMST")
td.data = td.data.coarsen(x=3, y=3, GMST=3, boundary="trim").reduce(np.mean)
/Users/jakobharteg/miniconda3/envs/toad312/lib/python3.12/site-packages/pyproj/network.py:59: UserWarning: pyproj unable to set PROJ database path.
_set_context_ca_bundle_path(ca_bundle_path)
td.compute_shifts("thk", method=ASDETECT(), overwrite=True)
INFO: New shifts variable thk_dts: min/mean/max=-1.000/-0.223/0.897 using 1642 grid cells. Skipped 58.6% grid cells: 0 NaN, 2327 constant.
The td.compute_clusters function accepts clustering methods from the sklearn.cluster module.
from sklearn.cluster import DBSCAN, HDBSCAN
# HDBSCAN
td.compute_clusters(
"thk",
method=HDBSCAN(
min_cluster_size=5,
),
shift_threshold=0.5,
)
# DBSCAN
td.compute_clusters(
"thk",
method=DBSCAN(
eps=0.1,
min_samples=5,
metric="euclidean", # optional, defaults to 'euclidean'
),
shift_threshold=0.5,
)
INFO: New cluster variable thk_dts_cluster: Identified 25 clusters in 1,633 pts; Left 23.1% as noise (378 pts).
INFO: New cluster variable thk_dts_cluster_1: Identified 28 clusters in 1,633 pts; Left 87.0% as noise (1,420 pts).
td
 TOAD Object
TOAD Object
Variable Hierarchy:
▶
base var thk
(1 shifts, 2 clusterings)
Hint: to access the xr.dataset call td.data
<xarray.Dataset> Size: 7MB
Dimensions: (GMST: 116, y: 63, x: 63)
Coordinates:
* GMST (GMST) float64 928B 0.0701 0.1901 0.3101 ... 13.75 13.87
* y (y) float64 504B -3e+06 -2.904e+06 ... 2.952e+06
* x (x) float64 504B -3e+06 -2.904e+06 ... 2.952e+06
Data variables:
thk (GMST, y, x) float32 2MB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0
thk_dts (GMST, y, x) float32 2MB nan nan nan nan ... nan nan nan
thk_dts_cluster (GMST, y, x) int32 2MB -1 -1 -1 -1 -1 ... -1 -1 -1 -1 -1
thk_dts_cluster_1 (GMST, y, x) int32 2MB -1 -1 -1 -1 -1 ... -1 -1 -1 -1 -1
Attributes: (12/13)
CDI: Climate Data Interface version 1.9.6 (http://mpimet.mpg.d...
proj4: +lon_0=0.0 +ellps=WGS84 +datum=WGS84 +lat_ts=-71.0 +proj=...
CDO: Climate Data Operators version 1.9.6 (http://mpimet.mpg.d...
source: PISM (development v1.0-535-gb3de48787 committed by Julius...
institution: PIK / Potsdam Institute for Climate Impact Research
author: Julius Garbe (julius.garbe@pik-potsdam.de)
... ...
title: garbe_2020_antarctica
Conventions: CF-1.9
projection: Polar Stereographic South (71S,0E)
ice_density: 910. kg m-3
NCO: netCDF Operators version 4.7.8 (Homepage = http://nco.sf....
Modifications: Modified by Jakob Harteg (jakob.harteg@pik-potsdam.de) Se...Defining your own clustering method¶
You can also define your own clustering method by extending the sklearn.base.ClusterMixin and sklearn.base.BaseEstimator classes:
from sklearn.base import ClusterMixin, BaseEstimator
# Your custom clustering class must inherit from ClusterMixin and BaseEstimator
class MyClusterer(ClusterMixin, BaseEstimator):
# Pass params to your method here
def __init__(self, my_param):
self.my_param = my_param
# required method to perform clustering
def fit_predict(self, X: np.ndarray, y=None, **kwargs):
# X = coords (time, x, y, z)
# y = weights / detection signal
cluster_labels_array = ... # your clustering algorithm here
return cluster_labels_array
# Then apply it with TOAD
# td.compute_clusters('thk',
# method=MyClusterer(
# my_param=(1, 2.0, 2.0), # time, x, y thresholds
# ),
# shift_threshold=0.8,
# overwrite=True,
# )
Real example of a custom clustering method:
from sklearn.base import BaseEstimator, ClusterMixin
class ExampleClusterer(ClusterMixin, BaseEstimator):
# required method
def __init__(self, my_param=(0.5, 1.0, 1.0)):
self.my_param = my_param
# required method
def fit_predict(self, X: np.ndarray, y=None, **kwargs):
# X = coords (time, x, y, z)
# y = weights / detection signal
# Perform extremely crude clustering
clusters = []
cluster_labels_array = []
for point in X:
for i, centroid in enumerate(clusters):
if all(abs(point - centroid) <= self.my_param):
break
else:
clusters.append(point)
i = len(clusters) - 1
cluster_labels_array.append(i)
return cluster_labels_array
td.compute_clusters(
"thk",
method=ExampleClusterer(
my_param=(0.5, 2.0, 2.0), # time, x, y thresholds
),
shift_threshold=0.9,
# overwrite=True,
)
INFO: New cluster variable thk_dts_cluster_2: Identified 17 clusters in 1,103 pts; Left 0.0% as noise (0 pts).
td
 TOAD Object
TOAD Object
Variable Hierarchy:
▶
base var thk
(1 shifts, 3 clusterings)
Hint: to access the xr.dataset call td.data
<xarray.Dataset> Size: 9MB
Dimensions: (GMST: 116, y: 63, x: 63)
Coordinates:
* GMST (GMST) float64 928B 0.0701 0.1901 0.3101 ... 13.75 13.87
* y (y) float64 504B -3e+06 -2.904e+06 ... 2.952e+06
* x (x) float64 504B -3e+06 -2.904e+06 ... 2.952e+06
Data variables:
thk (GMST, y, x) float32 2MB 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0
thk_dts (GMST, y, x) float32 2MB nan nan nan nan ... nan nan nan
thk_dts_cluster (GMST, y, x) int32 2MB -1 -1 -1 -1 -1 ... -1 -1 -1 -1 -1
thk_dts_cluster_1 (GMST, y, x) int32 2MB -1 -1 -1 -1 -1 ... -1 -1 -1 -1 -1
thk_dts_cluster_2 (GMST, y, x) int32 2MB -1 -1 -1 -1 -1 ... -1 -1 -1 -1 -1
Attributes: (12/13)
CDI: Climate Data Interface version 1.9.6 (http://mpimet.mpg.d...
proj4: +lon_0=0.0 +ellps=WGS84 +datum=WGS84 +lat_ts=-71.0 +proj=...
CDO: Climate Data Operators version 1.9.6 (http://mpimet.mpg.d...
source: PISM (development v1.0-535-gb3de48787 committed by Julius...
institution: PIK / Potsdam Institute for Climate Impact Research
author: Julius Garbe (julius.garbe@pik-potsdam.de)
... ...
title: garbe_2020_antarctica
Conventions: CF-1.9
projection: Polar Stereographic South (71S,0E)
ice_density: 910. kg m-3
NCO: netCDF Operators version 4.7.8 (Homepage = http://nco.sf....
Modifications: Modified by Jakob Harteg (jakob.harteg@pik-potsdam.de) Se...We can inspect all method params in the attributes
# get attributes of last cluster variable
td.get_clusters(td.cluster_vars[-1]).attrs
{'standard_name': 'land_ice_thickness',
'long_name': 'land ice thickness',
'units': 'm',
'pism_intent': 'model_state',
'time_dim': 'GMST',
'method_name': 'ExampleClusterer',
'toad_version': '0.3',
'base_variable': 'thk',
'variable_type': 'cluster',
'method_ignore_nan_warnings': 'False',
'method_lmin': '5',
'method_segmentation': 'two_sided',
'cluster_ids': array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16]),
'shift_threshold': 0.9,
'shift_selection': 'local',
'shift_direction': 'both',
'scaler': 'StandardScaler',
'time_weight': 1,
'n_data_points': 1103,
'runtime_preprocessing': 0.020125150680541992,
'runtime_clustering': 0.015676021575927734,
'runtime_total': 0.03580117225646973,
'shifts_variable': 'thk_dts',
'method_my_param': '(0.5, 2.0, 2.0)'}
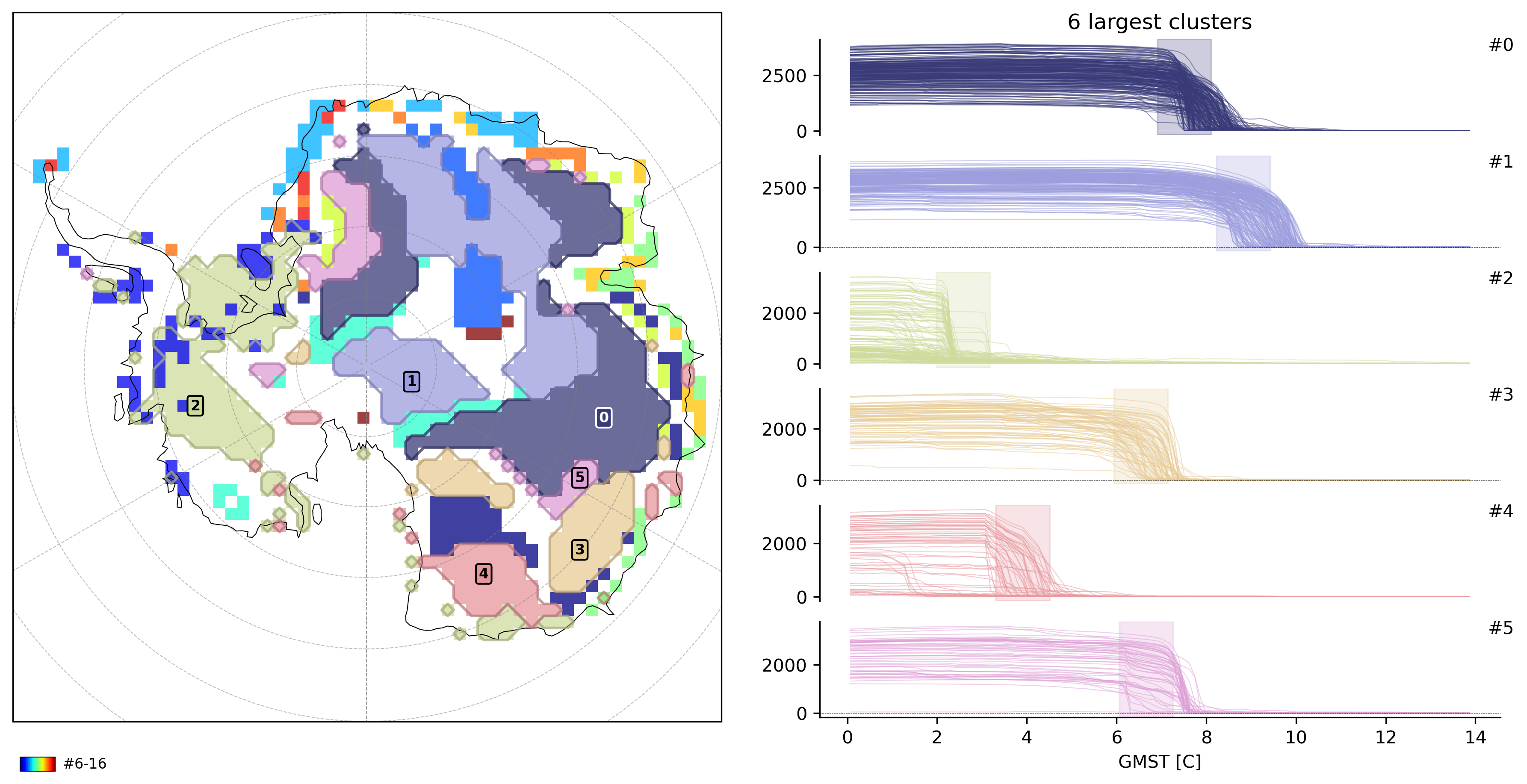
td.plot.overview(td.cluster_vars[-1], map_style={"projection": "south_pole"});